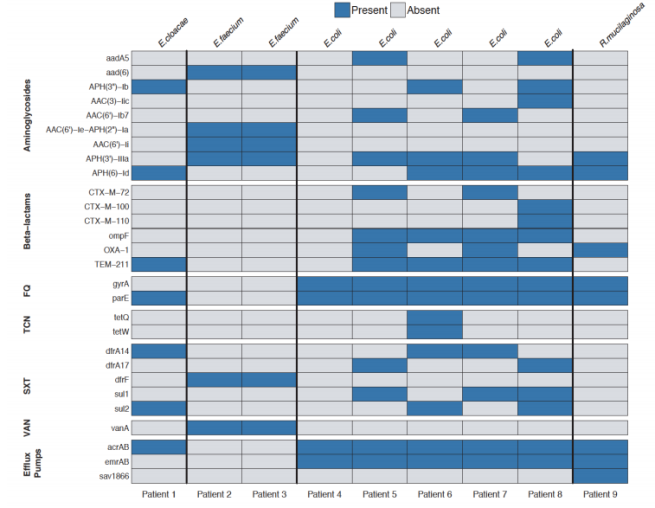
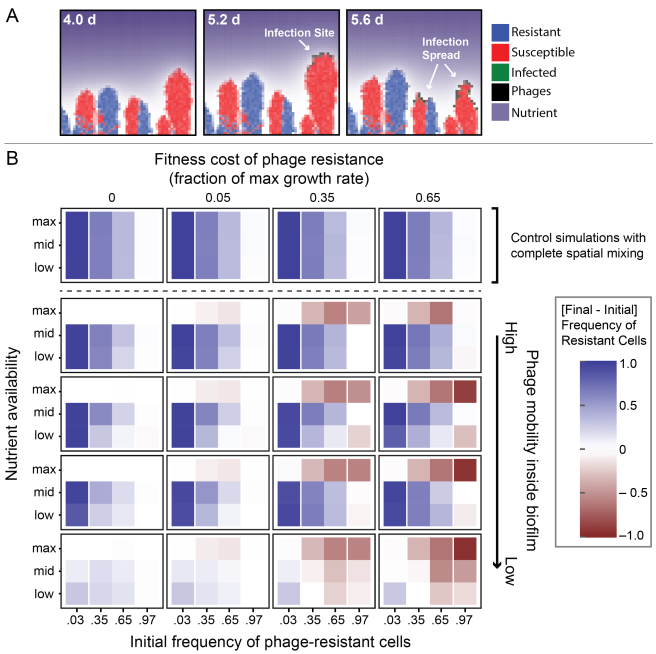
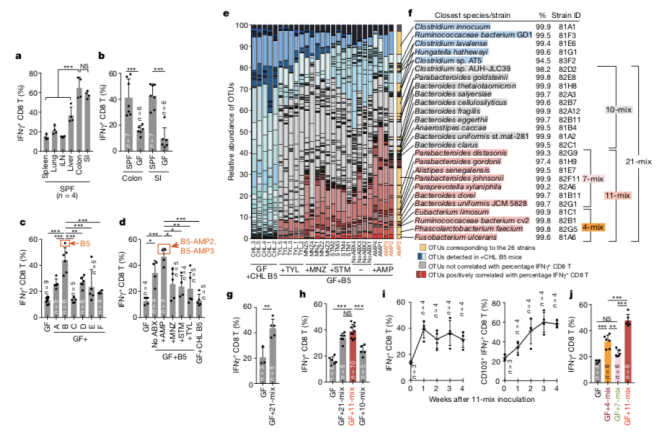
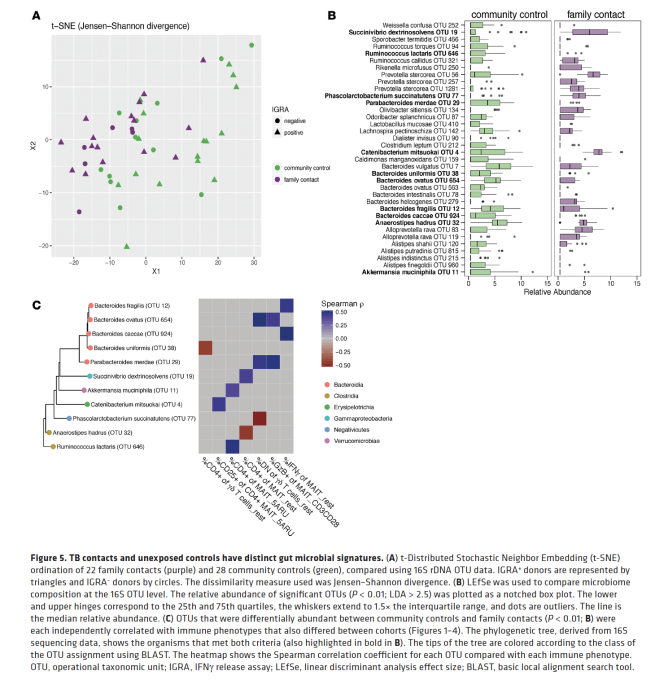
As a part of a newly awarded NIH grant in collaboration with the Gerber lab at Harvard Medical School/Brigham and Women’s Hospital, two PhD positions are available immediately in the Bucci lab at UMass Dartmouth to assist in development of computational models (and related software) for analyzing microbiome using advanced statistical and machine learning methods. In these positions, students will will have the opportunity to work on real, clinically relevant biomedical problems as well as help develop advanced machine learning methods to predict microbiome dynamics and their correlation with clinical outcome. There will also be an opportunity to publish and pursue independent research projects. These positions would be a good fit for very talented recent B.S. or M.S. graduate seeking to gain more research experience. Techniques used in the research, include Bayesian inference models, dynamical systems inference from sparse data, machine learning models, and approximate inference methods.
The Bucci lab (www.vannibucci.org) in the Bioengineering Department at UMass Dartmouth develops novel statistical/machine learning models and high-throughput experimental systems to understand the role of the microbiota in human diseases. A particular focus of the is using these computational methods to design probiotic assemblies that maximize certain host response including clearance pf pathogens or stimulation of anti-inflammatory conditions. We have applied these methods to a number of clinically relevant questions including understanding dynamic effects of antibiotics, infections and dietary changes on the microbiome, and designing bacteriotherapies for Clostridium difficile infection and inflammatory bowel disease.
Qualifications:
- Minimum of a Bachelor’s degree in physics, computer science, applied mathematics, statistics, or other highly quantitative discipline
- Strong Python development skills and experience implementing machine learning algorithms; TensorFlow experience desirable
- Formal coursework in algorithms, software design, machine learning, and probability/statistics required; you will need to understand quite a bit of theory
- Curiosity about biology/medical applications; microbiome experience not required
- Superior communication skills; you will be expected to contribute to writing scientific papers
Send cover letter and CV to vbucci@umassd.edu. Include a copy of your transcripts with grades. Applications without a cover letter specifically responsive to this posting cannot be considered.